How to implement internal dynamics of the head direction network in brain inspired 1D SLAM?
The excerpt note is from the Michael’s book. The example code is implemented by Fangwen, which can help us to understand the process and principle of the method.
Michael Milford. Robot Navigation from Nature: Simultaneous Localisation, Mapping, and Path Planning Based on Hippocampal Models. Springer-Verlag Berlin Heidelberg Press, pp. 68-69, 2008.
To achieve stable activity profiles in the head direction network, a system of localised excitation and global inhibition is used. The internal dynamics of the head direction cells are shown in Fig. 7.8.
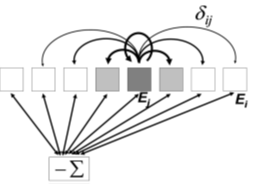
Fig. 7.8. Head direction cells excite themselves and other cells. The total activity sum is used to inhibit all cells equally.
Localised Excitation
Each cell is connected to all the cells in the network including itself via excitatory links. The self-connected weight is the largest, with weight values dropping off in a discrete Gaussian manner for connections to more distant cells. The matrix storing the excitatory weight values between all cells,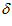 , is given by:
, is given by:
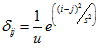
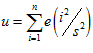
where is the variance of the Gaussian distribution used to form the weights profile.
is the variance of the Gaussian distribution used to form the weights profile.
During an experiment, activity in each of the head direction cells is projected through the appropriate excitatory links back into the head direction cells. The updated cell activity level after excitation,  , is given by:
, is given by:
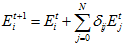
where 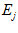 is the excitatory cell and
is the excitatory cell and 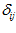 is the excitatory link value from cell
is the excitatory link value from cell  to cell
to cell  .
.
Global Inhibition
Global inhibition based on the total activity in the network is performed to ensure that without input from allothetic stimuli, smaller activity packets will gradually ‘die out’. The updated activity level of a cell after inhibition, 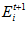 , is given by:
, is given by:
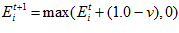
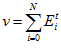
Global Normalisation
To ensure that the total activity sum in the head direction network stays constant, a global normalisation step is performed after the allothetic association, allothetic calibration, and ideothetic update steps. The cell activity levels after normalisation, 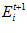 ,are given by:
,are given by:
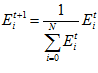
clear all;
global HDCELLS; % The hdcells network
HDCELLS = zeros(1,36);
% HDCELLS(15)=1;
HDCELLS(11)=0.05;
HDCELLS(12)=0.20;
HDCELLS(13)=0.25;
HDCELLS(14)=0.80;
HDCELLS(15)=0.28;
HDCELLS(16)=0.22;
HDCELLS(17)=0.16;
HDCELLS(18)=0.08;
global HD_DIMENSION_THETA; % The hdcell theta dimension
HD_DIMENSION_THETA = 36;
theta_size = 2*pi/HD_DIMENSION_THETA; % 0.6283 radian 36 deg
global HD_TH_SUM_SIN_LOOKUP;
global HD_TH_SUM_COS_LOOKUP;
HD_TH_SUM_SIN_LOOKUP = sin((1:HD_DIMENSION_THETA).*theta_size);
HD_TH_SUM_COS_LOOKUP = cos((1:HD_DIMENSION_THETA).*theta_size);
global HD_CELLS_TO_AVG;
global HD_AVG_TH_WRAP;
HD_CELLS_TO_AVG = 3;
HD_AVG_TH_WRAP = [(HD_DIMENSION_THETA - HD_CELLS_TO_AVG + 1):HD_DIMENSION_THETA 1:HD_DIMENSION_THETA 1:HD_CELLS_TO_AVG];
global HD_WEIGHT_EXCITATION_DIMENSION;
HD_WEIGHT_EXCITATION_DIMENSION = 7;
global HD_WEIGHT_INHIBITION_DIMENSION;
HD_WEIGHT_INHIBITION_DIMENSION = 5;
global HD_WEIGHT_EXCITATION_DIMENSION_HALF;
global HD_WEIGHT_INHIBITION_DIMENSION_HALF;
HD_WEIGHT_EXCITATION_DIMENSION_HALF = floor(HD_WEIGHT_EXCITATION_DIMENSION/2);
HD_WEIGHT_INHIBITION_DIMENSION_HALF = floor(HD_WEIGHT_INHIBITION_DIMENSION/2);
global HD_WEIGHT_EXCITATION_VARIANCE;
HD_WEIGHT_EXCITATION_VARIANCE = 2;
global HD_WEIGHT_INHIBITION_VARIANCE;
HD_WEIGHT_INHIBITION_VARIANCE =3;
global HD_GLOBAL_INHIBITION;
HD_GLOBAL_INHIBITION = 0.00002;
global HD_WEIGHT_EXCITATION; % the hdcell local excitation weight matricies
global HD_WEIGHT_INHIBITION; % the hdcell local inhibition weight matricies
HD_WEIGHT_EXCITATION = test_create_hdcell_weights(HD_WEIGHT_EXCITATION_DIMENSION,HD_WEIGHT_EXCITATION_VARIANCE);
HD_WEIGHT_INHIBITION = test_create_hdcell_weights(HD_WEIGHT_INHIBITION_DIMENSION,HD_WEIGHT_INHIBITION_VARIANCE);
%% Local Excitation
global HD_E_TH_WRAP;
HD_E_TH_WRAP = [(HD_DIMENSION_THETA - HD_WEIGHT_EXCITATION_DIMENSION_HALF + 1):HD_DIMENSION_THETA 1:HD_DIMENSION_THETA 1:HD_WEIGHT_EXCITATION_DIMENSION_HALF];
hdcells_le_new = zeros(1,HD_DIMENSION_THETA);
for z=1:HD_DIMENSION_THETA
if HDCELLS(z) ~= 0
hdcells_le_new(HD_E_TH_WRAP(z:z+HD_WEIGHT_EXCITATION_DIMENSION-1)) = ...
hdcells_le_new(HD_E_TH_WRAP(z:z+HD_WEIGHT_EXCITATION_DIMENSION-1))+ HDCELLS(z).*HD_WEIGHT_EXCITATION ;
end
end
HDCELLS = HDCELLS + hdcells_le_new;
global HD_I_TH_WRAP;
HD_I_TH_WRAP = [(HD_DIMENSION_THETA - HD_WEIGHT_INHIBITION_DIMENSION_HALF + 1):HD_DIMENSION_THETA 1:HD_DIMENSION_THETA 1:HD_WEIGHT_INHIBITION_DIMENSION_HALF];
%% local inhibition
hdcells_li_new = zeros(1,HD_DIMENSION_THETA);
for z=1:HD_DIMENSION_THETA
if HDCELLS(z) ~= 0
hdcells_li_new(HD_I_TH_WRAP(z:z+HD_WEIGHT_INHIBITION_DIMENSION-1)) = ...
hdcells_li_new(HD_I_TH_WRAP(z:z+HD_WEIGHT_INHIBITION_DIMENSION-1))+ HDCELLS(z).*HD_WEIGHT_INHIBITION ;
end
end
HDCELLS = HDCELLS - hdcells_li_new;
%% global inhibition
for i=1 : HD_DIMENSION_THETA
if HDCELLS(i) >= HD_GLOBAL_INHIBITION
HDCELLS(i) = HDCELLS(i) - HD_GLOBAL_INHIBITION;
else
HDCELLS(i) = 0;
end
end
%% Global Normalisation
total = 0;
total = sum(HDCELLS);
HDCELLS = HDCELLS./total;
%% Get the current head direction
[value, z] = max(HDCELLS);
z_HDCELLS = zeros(1,HD_DIMENSION_THETA);
z_HDCELLS(HD_AVG_TH_WRAP(z:z+HD_CELLS_TO_AVG*2)) = HDCELLS(HD_AVG_TH_WRAP(z:z+HD_CELLS_TO_AVG*2));
out_theta = mod(atan2(sum(HD_TH_SUM_SIN_LOOKUP *(z_HDCELLS')), sum(HD_TH_SUM_COS_LOOKUP *(z_HDCELLS')))/theta_size, HD_DIMENSION_THETA);
%% create local excitation weight or local inhibition weight
function [weight] = test_create_hdcell_weights(dim,var)
dim_centre = floor(dim/2) + 1;
weight=zeros(1,dim);
for z=1:dim
weight(z) = 1/(var*sqrt(2*pi))*exp(-(z-dim_centre)^2)/(2*var^2);
end
total = sum(weight);
weight = weight./total;
end
About
CogNav Blog
New discovery worth spreading on cognitive navigation in neurorobotics and neuroscience
Recent Posts
- How to build a bio-inspired hardware implementation of an analog spike-based hippocampus memory model?
- How does the brain select what to remember during sleep?
- How hippocampal activity encodes numerous memories of specific events in life?
- How egocentric coding properties arise from its presynaptic inputs, and how egocentric cells represent items across different behavioral contexts?
- How the medial entorhinal cortex develops during learning and influences memory?
Tags
Categories
- 3D Movement
- 3D Navigation
- 3D Path Integration
- 3D Perception
- 3D SLAM
- 3D Spatial Representation
- AI Navigation
- Bio-Inspired Robotics
- Brain-Inspired Navigation
- Cognitive Map
- Cognitive Navigation
- Episodic Memory
- Excerpt Notes
- Flying Vehicle Navigation
- Goal Representation
- Insect Navigation
- Learning to Navigate
- Neural Basis of Navigation
- Path Integration
- Path Planning
- Project
- Research Tips
- Robotic Vision
- Self-Flying Vehicles
- Spatial Cognition
- Spatial Cognitive Computing
- Spatial Coordinate System
- Spatial Memory
- Time
- Unclassified
- Visual Cortex
- Visual Cue Cells
Links
- Laboratory of Nachum Ulanovsky
- Jeffery Lab
- BatLab
- The NeuroBat Lab
- Taube Lab
- Laurens Group
- Romani Lab
- Moser Group
- O’Keefe Group
- DoellerLab
- MilfordRobotics Group
- The Space and Memory group
- Angelaki Lab
- Spatial Cognition Lab
- McNaughton Lab
- Conradt Group
- The Fiete Lab
- The Cacucci Lab
- The Burak Lab
- Knierim Lab
- Clark Spatial Navigation & Memory Lab
- Computational Memory Lab
- The Dombeck Lab
- Zugaro Lab
- Insect Robotics Group
- The Nagel Lab
- Basu Lab
- Spatial Perception and Memory lab
- The Neuroecology lab
- The Nagel Lab
- Neural Modeling and Interface Lab
- Memory and Navigation Circuits Group
- Neural Circuits and Memory Lab
- The lab of Arseny Finkelstein
- The Epstein Lab
- Gu Lab (Spatial Navigation and Memory)
- Fisher Lab (Neural Circuits for Navigation)
- The Alexander Lab (Spatial Cognition and Memory)
- Harvey Lab (Neural Circuits for Navigation)
- Buzsáki Lab
- ……
